How EPITOPIC’s antibody epitope fingerprinting works
EPITOPIC has developed unique technologies to overcome common problems associated with selection from biological libraries by excluding the unwanted biological effects of recent selection procedures. Unnecessary selection steps are avoided and new tools allow the identification without tremendous enrichment.

Our specially designed naïve library covers approximately five billion different random 16-mer peptide sequences, covering most 6-mer amino acid combinations. Due to the special design and care in the generation, the peptide phage library does not contain overrepresented single sequences, even when the library is replicated. This enables the success of epitope fingerprinting, which fails when other starting libraries are used.
Only minimal amounts of antibodies (µg) or serum (ca 100 µl) are required. Theoretically, the selection and enrichment of binders requires not more than a few hundred antibody molecules, so even the epitopes of antibodies in a serum can be identified.
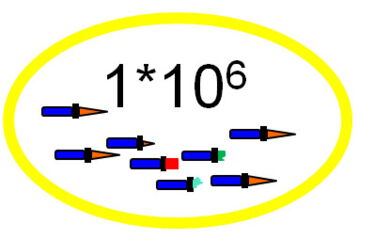
For each epitope up to thousand variants of 16-mer sequences resembling the epitope motif are enriched in a single step, but they are still hidden in up to one million different sequences. Due to the statistically even amino acid distribution even a minimal enrichment is sufficient for the following steps.
NGS is error prone. Using several specially developed algorithms, we can identify sequencing errors. In our set up with illumina sequencers, up to 30% of the obtained single sequences are expected to contain errors and are removed prior to the subsequent analysis. This avoids unnecessary and expensive oversampling, which would multiply the project costs.
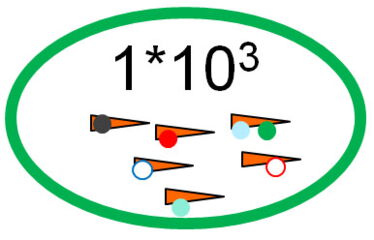
Datasets with up to a million peptides are generated by translating the peptide genes. They contain sometimes only a few hundred enriched sequences with epitope motifs. This is sufficient because enrichment will not be detected based on single sequences, but statistical analysis is used to identify a pool of similar sequences sharing enriched motifs.

Statistical fishing for epitope sequences from these large datasets is performed with the help of proprietary software that allows the identification of enriched continuous and discontinuous peptide motifs resembling the antigen or sharing sequence features. The surrounding amino acids of 16-mer peptides often support the special peptide epitope conformations required for binding to the antibody.
Epitope fingerprints obtained combining the information from many peptides allow to understand not only the location of the epitope but also individual preferences of antibodies with respect to surrounding or intermitting amino acid residues. Analysis of aligned sequences helps predict specificity with respect to bound proteins in comparison to related or non-related proteins from the host proteome or other species. This is unique, and no additional experiments are required.